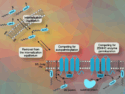
A promising approach to new antiviral drugs involves the use of small interfering ribonucleic acids (siRNAs) that bind to complementary viral RNA. They induce degradation and block viral replication. Such siRNA drugs could be rapidly adjusted to emerging virus variants. However, it is still challenging to achieve efficient cell-specific delivery of siRNA therapeutics.
Thomas Carell, Ludwig Maximilian University of Munich, Germany, and colleagues have designed siRNAs that are complementary to different viral RNA target regions of SARS-CoV-2. The siRNAs were prepared using an automated RNA synthesizer. The siRNAs were chemically stabilized by partially modifying the sugar-phosphate backbone with methoxy substituents.
The researchers also introduced an alkyne unit into the siRNA (schematically pictured below), which allowed modification of the siRNA with ligands such as peptides using Cu(I)-catalyzed click chemistry. The siRNA was conjugated to a small peptide that binds to the angiotensin-converting enzyme 2 (ACE2) receptor, which is important for the cell entry of SARS-CoV-2.
The siRNA-peptide conjugate was able to reduce virus replication and virus-induced cell death in model lung tissues. This shows the potential of targeted siRNAs as antiviral therapeutics. The adjustment of siRNA sequences could allow for a fast adaptation of the antiviral activity to different variants of concern.
- Suppression of SARS‐CoV‐2 Replication with Stabilized and Click‐Chemistry Modified siRNAs,
Franziska R. Traube, Marcel Stern, Annika J. Tölke, Martina Rudelius, Ernesto Mejías-Pérez, Nada Raddaoui, Beate M. Kümmerer, Céline Douat, Filipp Streshnev, Manuel Albanese, Paul R. Wratil, Yasmin V. Gärtner, Milda Nainytė, Grazia Giorgio, Stylianos Michalakis, Sabine Schneider, Hendrick Streeck, Markus Müller, Oliver T. Keppler, Thomas Carell,
Angew. Chem. Int. Ed. 2022.
https://doi.org/10.1002/anie.202204556
Also of Interest
- Collection: SARS-CoV-2 Virus
What we know about the new coronavirus and COVID-19