Probing DNA mismatches is useful for understanding DNA mismatch repair (MMR), a critical process in maintaining genomic stability through fixing various forms of errors and damages in DNA. Current strategies rely on the insertion of molecular probes into DNA, which still lacks selectivity and may disrupt normal base pairs.
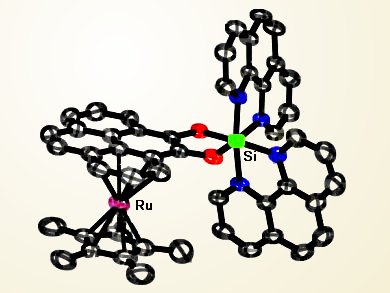
In an effort towards selective DNA mismatch recognition, Eric Meggers and co-workers, Philipps-Universität Marburg, Germany, and Xiamen University, China, have developed a novel dinuclear complex, featuring an octahedral Si(IV) core with a peripheral pyrene ligand sandwiching Ru(II). This complex can selectively stabilize and thus detect cytosine−cytosine and cytosine−thymine mismatches in DNA duplexes due to the specific π−stacking between the entire Ru sandwich unit and mismatched sites.
This research represents the first example of an insertion−intercalation binding mode between a probe and DNA, which can further shed light on the design of selective and accurate probes for mapping DNA mismatches.
- DNA Mismatch Recognition by a Hexacoordinate Silicon Sandwich−Ruthenium Hybrid Complex,
C. Fu, K. Harms, L. Zhang, E. Meggers
Organometallics 2014, 33, 3219–3222.
DOI: 10.1021/om500367a