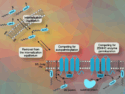
Viruses such as SARS-CoV-2, which causes COVID-19, can synthesize multiple proteins as one unit. Viral enzymes called proteases then recognize specific amino-acid sequences in these polyproteins and cut them to release the individual proteins. The SARS-CoV-2 papain-like protease (PLpro), for example, plays an essential role in processing viral proteins needed for replication. However, some human proteins also contain the sequences that are cut by these enzymes (known as short stretches of homologous host-pathogen sequences, or SSHHPS). Some of these human protein targets are involved in generating an immune response.
Patricia M. Legler, U.S. Naval Research Laboratory, Washington, DC, USA, and colleagues have found other targets of PLpro in the human proteome, including proteins involved in cardiovascular function, blood clotting, and inflammation. The team aimed to comprehensively identify human proteins that contain SSHHPS, examine their functions, and check whether PLpro can cleave them in vitro. The researchers used a program called PHI-BLAST (Pattern-Hit Initiated Basic Local Alignment Search Tool) to search a database of known human proteins for sequences similar or identical to the SARS-CoV-2 SSHHPS.
The team found potential target proteins that had cardiovascular, inflammatory, kidney, respiratory, or blood-related functions, suggesting a link between the inactivation of these proteins and COVID-19 symptoms. For example, two of the proteins containing SSHHPS were cardiac myosins (proteins that play a role in heart muscle contraction), one was an anti-coagulant, and another was an anti-inflammatory protein. Inactivation of these proteins by PLpro is consistent with symptoms such as heart damage, blood clots, and inflammation. The team confirmed that PLpro could cut these protein sequences in vitro.
The team performed the same analysis on SSHHPS for the protease of the Zika virus and found possible target proteins associated with neurological development and neurological disorders, consistent with the effects of Zika. According to the researchers, these results suggest a correlation between the symptoms caused by viruses and their genomic sequences. This might be useful for predictive applications.
- The SARS-CoV-2 SSHHPS Recognized by the Papain-like Protease,
Nathanael D. Reynolds, Nathalie M. Aceves, Jinny L. Liu, Jaimee R. Compton, Dagmar H. Leary, Brendan T. Freitas, Scott D. Pegan, Katarina Z. Doctor, Fred Y. Wu, Xin Hu, Patricia M. Legler,
ACS Infect. Dis. 2021.
https://doi.org/10.1021/acsinfecdis.0c00866
Also of Interest
- Collection: SARS-CoV-2 Virus
What we know about the new coronavirus and COVID-19