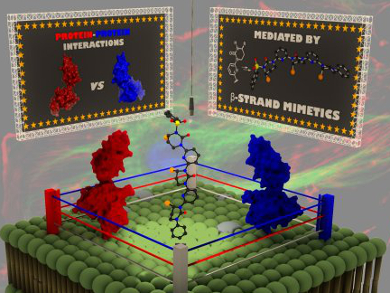
In the fight against disease, classical strategies typically target localized and highly specific interactions between enzymes and their substrates. An alternative strategy is to look at protein–protein interactions that occur between extremely large and relatively featureless surfaces. A promising strategy for the recognition of such surfaces is the mimicry of secondary structural protein elements. While the mimicry of helical structures has seen some success, reports on the mimicry of ß-sheet and ß-strand structures are rare.
By using a modular, iterative strategy, Sam Thompson, Andrew D. Hamilton and their co-workers, University of Oxford, UK, have designed pre-formed monomers bearing an amino acid side-chain that can undergo rapid assembly to give peptidomimetics. The resultant oligomers, which contain alternating pyridyl and six-membered cyclic ureas, accurately reproduce a recognition domain of several amino acid residues of a ß-strand, as shown by X-ray crystallography, NMR spectroscopy and molecular mechanics (MM) calculations.
- A Modular Synthesis of Conformationally Preorganised Extended β-Strand Peptidomimetics,
Tohru Yamashita, Peter C. Knipe, Nathalie Busschaert, Sam Thompson, Andrew D. Hamilton,
Chem Eur. J. 2015.
DOI: 10.1002/chem.201501366