Chemically altered nucleosides have found various applications ranging from chemical biology to nanotechnology. A common synthetic approach for modified nucleic acids is the automated solid-phase synthesis of oligonucleotides. However, the nature of these modifications is limited and often depends on their resilience to the harsh conditions imposed by solid-phase synthesis. Moreover, phosphoramidite building blocks, whether chemically modified or not, are incompatible with enzymatic amplification techniques, such as the polymerase chain reaction (PCR), and thus cannot be applied in many in vitro selection methods. The enzymatic polymerization of modified nucleoside triphosphates (dNTPs) is an alternative, milder, and more versatile way to generate chemically altered nucleic acids, yet is still relatively unexplored.
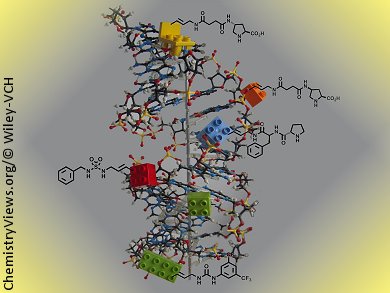
Marcel Hollenstein, University of Bern, Switzerland, has synthesized five base-modified nucleoside triphosphates adorned with functionalities that play a central role in organocatalysis (i.e., proline, urea, and sulfamide groups). These dNTPs are compatible with in vitro selection methods and could supplement the chemical collection of DNA enzymes when used in selection experiments. These modified dNTPs were found to be excellent substrates for three different DNA polymerases in various primer-extension reactions. Moreover, four were found to be efficient in PCR amplifications.
In addition, modified single stranded DNAs (ssDNAs) resulting from primer-extension reactions could serve as templates in PCR experiments and be converted into natural DNA amplicons. Thermal denaturation experiments showed that the modified nucleotides had a marginal impact on the stability of the duplexes, hinting at good acceptance of these side chains in the major groove.
- Synthesis of Deoxynucleoside Triphosphates that Include Proline, Urea, or Sulfonamide Groups and their Polymerase Incorporation into DNA,
M. Hollenstein,
Chem. Eur. J. 2012.
DOI: 10.1002/chem.201201662