In some proteins, the polypeptide chains wrap around themselves with occasional entanglements and/or covalent crosslinks (for example, disulfide-, ester-, or isopeptide bonds) upon folding, and thus, form topologies such as knots or slipknots. The discovery of these natural “topological proteins” has inspired researchers to develop artificial topological proteins. However, it is hard to control the chain entanglement and coupling sites in the biosynthesis process of proteins. Catenanes are examples of such entangled structures, where two or more rings are mechanically interlocked.
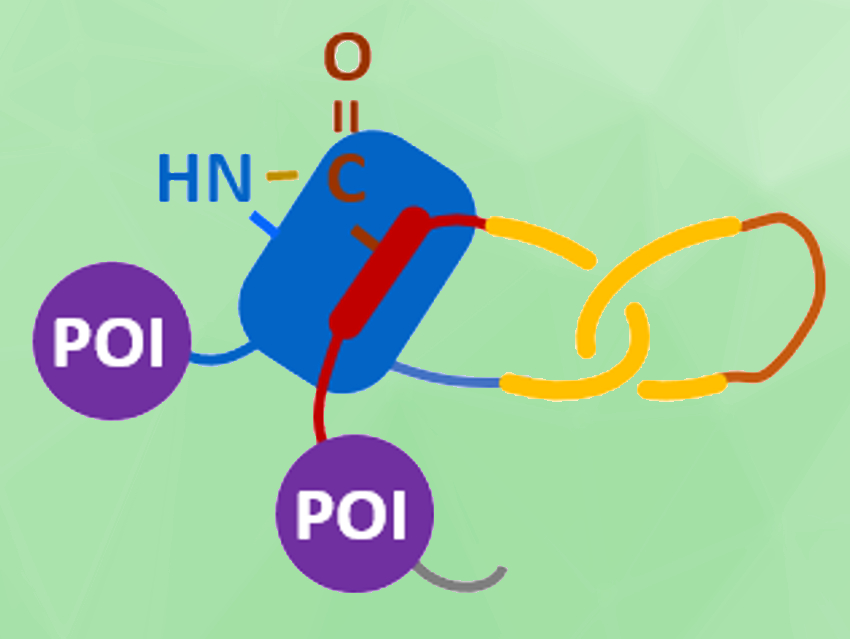
Junyu Xiao, Wen‐Bin Zhang, Peking University, Beijing, China, and colleagues have achieved a biosynthesis of protein heterocatenanes (pictured). The team used a programmed sequence of multiple processing steps, including chain entanglement, backbone cleavage, and spontaneous cyclization. They first designed a gene sequence (pictured below), called SpyCatcher-p53dim-SpyTag-TVMV-IntC1-p53dim-IntN1, where TVMV stands for the proteolytic site of tobacco vein mottling virus protease, and IntC1/IntN1 for the split-intein pair.
All components encoded in this gene sequence then play a role in the assembly process after the translation from gene to polypeptide: The SpyTag–SpyCatcher reaction is a peptide–protein coupling tool. The p53 dimerization domain (p53dim) can direct chain entanglement to provide a template for the synthesis of protein catenanes. The TVMV protease enables in–situ chain cleavage. The split-intein pair can be used for cyclizing proteins.
The resulting assembly process is pictured below. The protein chain first folds into an intramolecularly entangled structure. TVMV protease causes a chain cleavage at the loop region. Finally, the SpyTag–SpyCatcher reaction (pictured in blue/red) and the reaction between the slit-intein pair (pictured in lavender) close the first and second ring, respectively, to form the desired heterocatenane. Genes for additional proteins of interest (POIs, pictured in violet) can be introduced into the sequence to create branched heterocatenanes.
The heterocatenane’s topology was confirmed using SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis), LC–MS (liquid chromatography–mass spectrometry), controlled proteolytic digestion, and protein X-ray crystallography. The team’s modular approach for the biosynthesis of protein heterocatenanes converts proteins from one-chain to multi-chain molecules with higher complexity. It provides topological proteins so their structure-property relationships can be explored further.

- Cellular Synthesis and Crystal Structure of a Designed Protein Heterocatenane,
Yajie Liu, Zelin Duan, Jing Fang, Fan Zhang, Junyu Xiao, Wenbin Zhang,
Angew. Chem. Int. Ed. 2020.
https://doi.org/10.1002/anie.202005490