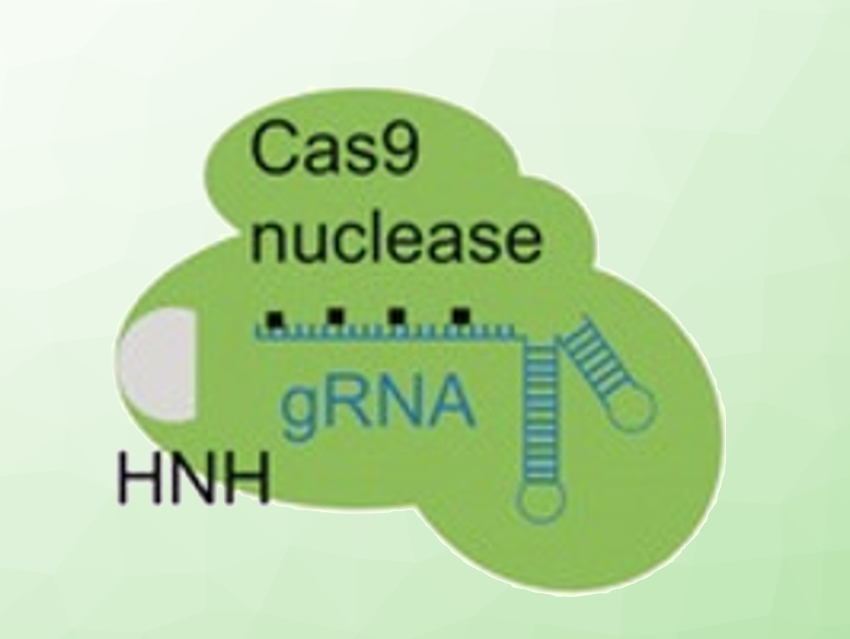
CRISPR-Cas9 is a useful genetic editing tool that allows for efficient and precise editing. It consists of guide RNA (gRNA), which targets specific sites in a DNA strand, and a Cas9 enzyme, which cuts the DNA at these sites. Controlling this technique by activating it at a certain time or at a particular location would be useful. Approaches to achieve this control have been mostly confined to engineering the Cas9 protein itself. However, protein engineering can be challenging.
Alexander Deiters, University of Pittsburgh, PA, USA, and colleagues have developed a light-activated CRISPR-Cas9 system by altering the gRNA that directs the Cas9 protein to the target editing site. This was achieved by replacing several of the nucleotides in the target-binding region of the gRNA with “photocaged” nucleotides. The team used 6‐nitropiperonyloxymethylene (NPOM)‐caged nucleobases. These chemical cages can be removed by light exposure (pictured below) to activate the system. The chemically modified gRNA can be prepared using standard solid-phase oligonucleotide synthesis.
The Cas9 protein and the caged gRNA were introduced into mammalian cells or zebrafish embryos. Light exposure activated gene editing with spatial and temporal control, both in cell cultures and in developing embryos. The team was able to edit different genomic loci, which shows the system’s universal applicability.

- Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light‐Activated Guide RNA,
Wenyuan Zhou, Wes Brown, Anirban Bardhan, Michael Delaney, Amber Ilk, Randy Rauen, Shoeb Kahn, Michael Tsang, Alexander Deiters,
Angew. Chem. Int. Ed. 2020.
https://doi.org/10.1002/anie.201914575