The oxidation of cysteine residues in proteins with glutathione (S-glutathionylation) in the presence of hydrogen peroxide controls many complex biological processes in cells. Therefore, the identification of specific target proteins susceptible to S-glutathionylation is important to understand the biological consequences of oxidative conditions. However, methods to identify S-glutathionylation at the proteomic level are scarce.
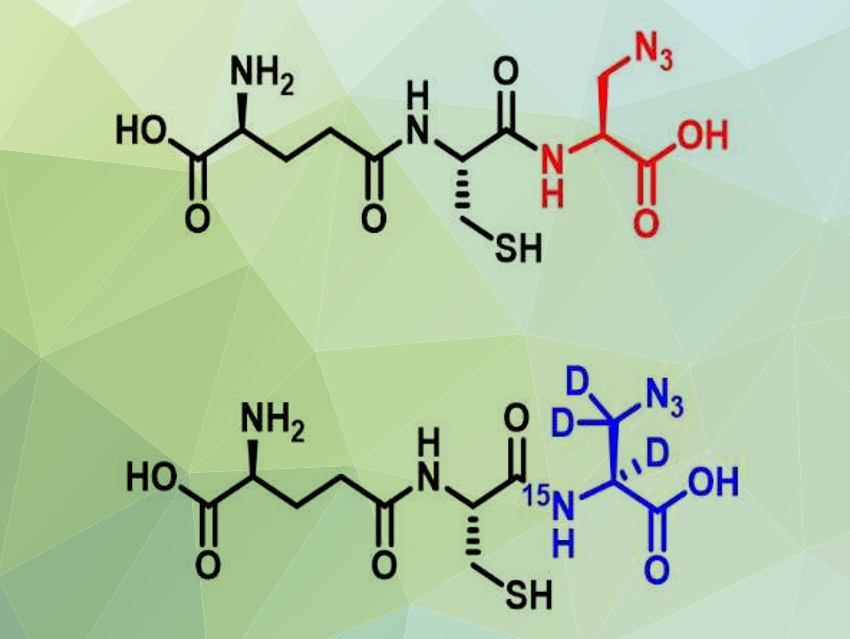
Young-Hoon Ahn, Wayne State University, Detroit, MI, USA, and colleagues have developed a mass-spectrometric method to identify and quantify target proteins of S-glutathionylation. The team used cells with a genetically engineered glutathione synthetase enzyme to make isotopically labeled glutathione derivatives with a “clickable” azide functional group (pictured). These two otherwise identical glutathione derivatives have a small mass difference. A click reaction with an alkyne-functionalized reagent enabled the team to label the glutathionylated proteins and to separate and identify them.
The researchers used this approach to find proteins that are susceptible to S-glutathionylation in cardiomyocytes (heart muscle cells). The team used two different batches of the cells and reacted them with either the heavy or the light glutathione derivative. The glutathionylation was induced by the addition of H2O2 and the modified peptides were labeled and extracted. Then, using mass spectrometry (MS), the team looked for pairs of peptides with exactly the same mass difference as the two glutathione derivatives and quantified them. They found elevated levels of S-glutathionylation on 797 cysteines. According to the team, this method could be broadly applicable to various biological systems to identify S-glutathionylation.
- Isotopically-labelled Clickable Glutathione to Quantify Protein S-Glutathionylation,
Garrett C. VanHecke, Maheeshi Yapa Abeywardana, Bo Huang, Young-Hoon Ahn,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900528