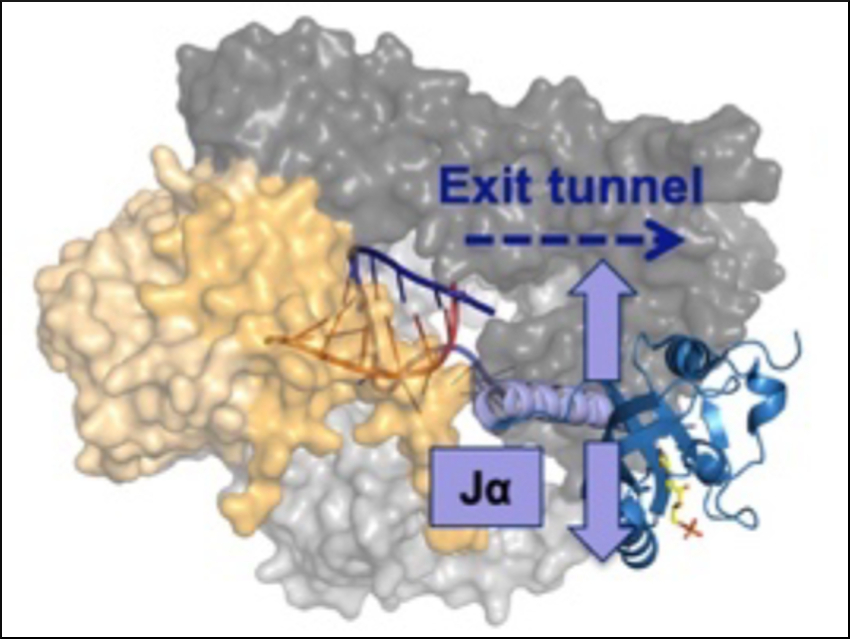
Natural photoresponsive proteins alter their shape when they are hit by sunlight. This can, for example, change the activity of an enzyme linked to the protein. Because light is a signal that can be applied with high precision, such proteins have become increasingly important for controlling the activity of enzymes in biochemical and medicinal research.
Susanne Brakmann and colleagues, Technische Universität Dortmund, Germany, have investigated how light-sensing “light-oxygen-voltage” (LOV) protein domains can be combined with a nucleic acid polymerase, an enzyme that is involved in the transcription of DNA to RNA. The team used bacteriophage T7 RNA polymerase (T7 RNAP, pictured in yellow) as a model enzyme. The light-switching of LOV is based on the unfolding of a helical part of the protein called Jα (pictured in lavender) upon excitation with light.
The team found that it is possible to covalently insert LOV domains at various positions of T7 RNAP while preserving its enzymatic activity. The activities of the LOV-polymerase fusion proteins are tunable by light. Depending on the exact position of LOV-insertion, the activities of the polymerase variants can be either increased or decreased by illumination with blue light (470 nm) (pictured below). These new polymerase fusion proteins could be useful as tools for studying the role of RNA and DNA in cellular, developmental, and disease-related processes.
.jpg)
- Optical control of transcription—genetically encoded photoswitchable variants of T7 RNA polymerase,
Swantje Seifert, Christiane Ehrt, Lena Lükfeldt, Melissa Lubeck, Frederik Schramm, Susanne Brakmann,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900298