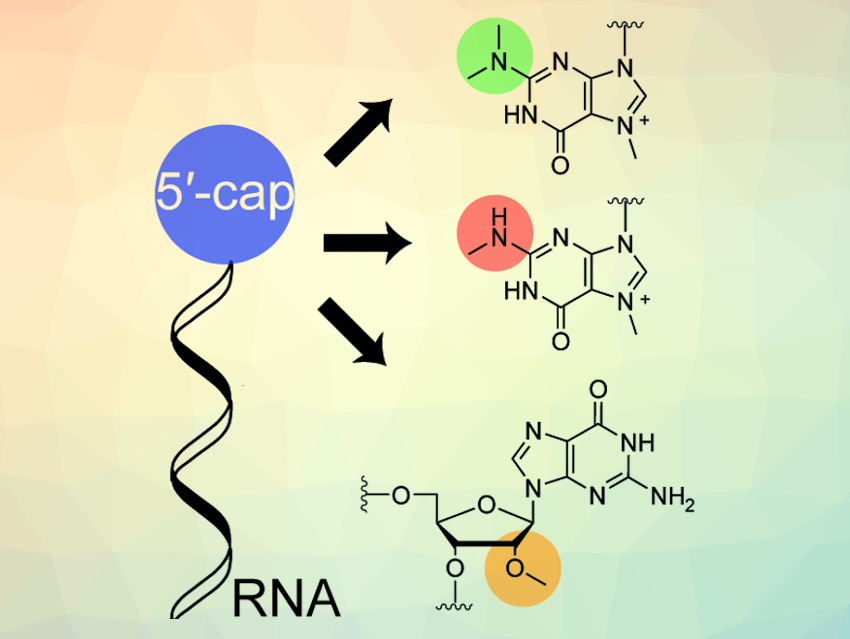
The RNA 5′-cap is an altered nucleotide on one end of some of the RNA transcripts produced in eukaryotic organisms. It is known to be heavily processed both during and after the transcription of DNA to RNA. It contains modifications such as enzymatic methylations at different sites within the 5′-cap (pictured in green, red, and orange above). However, the function and potential dynamics of some of these modifications have not yet been fully investigated. A way to synthesize these RNAs could help with this.
Francoise Debart, University of Montpellier, France, Andrea Rentmeister, University of Münster, Germany, and colleagues have combined chemical and enzymatic synthesis with site-specific methyltransferases to obtain highly cap-modified RNAs. The RNA substrates were prepared either by chemical synthesis or enzymatically.
Methyltransferases called GlaTgs and hTgs were used to obtain RNAs that are either mono- or di-methylated at the N2-position of guanine. This modification is commonly found in caps of snRNAs (small nuclear RNAs). The methyltransferase CMTR1 allowed access to the so-called cap1 structure, a structural motif in mRNA (messenger RNA) important for efficient translation.According to the researchers, this approach could allow further studies of RNA modification dynamics.
- Combining chemical synthesis and enzymatic methylation to access short RNAs with various 5′ caps,
Nils Muthmann, Théo Guez, Jean-Jacques Vasseur, Samie R Jaffrey, Francoise Debart, Andrea Rentmeister,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900037