Natural product research can profit from the advance of genomics technology. Usually, organisms are screened for natural products and, inevitably, known compounds are rediscovered in the process. Instead, thousands of microbial genomes can be quickly analyzed for gene clusters that might encode enzymes which synthesize new compounds. In addition, a genetic origin could be assigned to previously discovered compounds with no known biosynthesis.
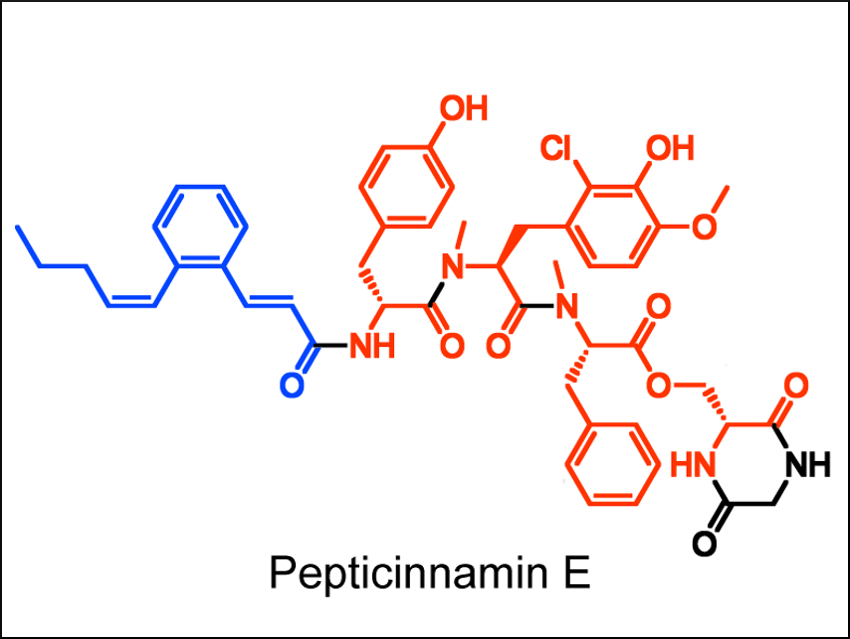
Bo Li and colleagues, University of North Carolina at Chapel Hill, USA, have used genomics tools to identify a biosynthetic origin for pepticinnamin E (pictured), a compound which acts as an inhibitor of the enzyme farnesyl transferase. Pepticinnamin E has potential applications in treating cancer and malaria. The compound also contains chemical motifs rarely observed in other natural products, e.g., a rare N-terminal cinnamoyl group (pictured in blue).
The team used this signature cinnamoyl moiety as a biosynthetic “hook”. The researchers looked for gene clusters that might encode enzymes for the synthesis of this group in a microbial genome database. They identified a gene cluster (pcm) which is responsible for the biosynthesis of this natural product. This gene cluster was found in a new producer of pepticinnamin E, Actinobacteria bacterium OK006. According to the researchers, this work showcases the power of genome mining and could help with further investigations of the biosynthesis of pepticinnamins.
- Targeted Rediscovery and Biosynthesis of the Farnesyl-Transferase Inhibitor Pepticinnamin E,
Kevin C. Santa Maria, Andrew N. Chan, Erinn M. O’Neill, Bo Li,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900025