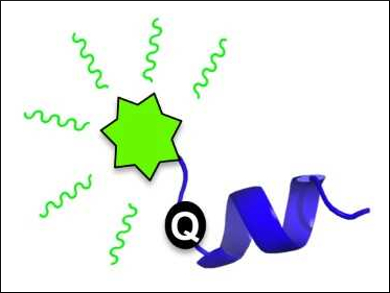
Many proteins are intrinsically disordered, meaning they lack a well-defined 3D structure. However, they often form a 3D structure upon interaction with a binding partner. Intrinsically disordered proteins and protein regions (IDPs and IDRs) are, thus, dynamic and can function as “signaling hubs” in protein interaction networks. However, observing their structural transitions can be challenging because the commonly used methods are low-throughput and require large sample quantities.
Amanda L. Garner and colleagues, University of Michigan, Ann Arbor, USA, have developed a fluorescence strategy for monitoring such structural transitions. The team studied the protein 4E-BP1, an intrinsically disordered protein that forms an α-helix upon binding to its ligand eIF4E. The fluorescence of a fluorescein-labeled 4E-BP1 peptide with a thiourea group is quenched in its disordered state by photoinduced electron transfer (PET, pictured below on the left).
When the &alpha:-helix forms, an increase in fluorescence is observed. This is due to the thiourea moving out of PET “quenching distance” (pictured below on the right). This detection of peptide structure works at low concentrations and can be used in high-throughput screenings. This finding could help to discover ligands that alter the secondary structure of peptides.

- A Conditionally Fluorescent Peptide Reporter of Secondary Structure Modulation,
Amanda Lee Garner, Oleta T. Johnson, Tanpreet Kaur,
ChemBioChem 2018.
https://doi.org/10.1002/cbic.201800377