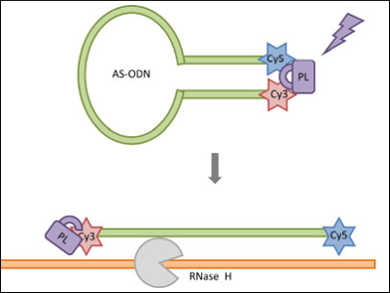
Light-activated (“caged”) oligonucleotides are useful molecules for investigating gene function and modulating gene expression with precise control. Cyclization is considered an efficient caging strategy, however, efficient strategies are still needed for caging larger biomolecules such as oligonucleotides, proteins, and polysaccharides.
Ivan J. Dmochowski, University of Pennsylvania, Philadelphia, USA, and colleagues have developed an efficient route for generating circular oligonucleotides through an intramolecular copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC). The cyclization step proceeds almost quantitatively without requiring further purification and was applied to different sequences. The resulting antisense oligodeoxynucleotides (AS-ODNs, pictured top) can be activated with light and bind to a target mRNA strand. The mRNA is then degraded by an enzyme, which prevents its translation (pictured bottom).
One sequence targeting the gfap gene achieved a tenfold reduction of GFAP protein expression after light activation. Circular oligonucleotides of this type should be readily adaptable for other targets, such as microRNA or long non-coding RNA (lncRNA).
- Efficient synthesis of light-triggered circular antisense oligonucleotides targeting cellular protein expression,
Linlin Yang, Hyun-Bum Kim, Jai-Yoon Sul, Sean B. Yeldell, James H. Eberwine, Ivan J. Dmochowski,
ChemBioChem 2018.
https://doi.org/10.1002/cbic.201800012