The quest for the de novo synthesis of life has focused on several key aspects: replication, compartmentalization, and metabolism. Many subsystems have been reported that feature one of these characteristics, but coupling these subsystems remains challenging.
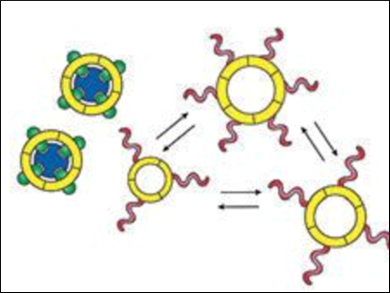
Sijbren Otto, University of Groningen, The Netherlands, and colleagues have developed a self-replication subsystem by designing molecules that can catalyze their own formation. The team showed how replication can be triggered by an effector molecule. They designed replication and effector-recognition subsystems and connected these by reversible covalent bonds (see scheme).

Mixing of the building blocks of the two subsystems produces a complex molecular network where self-replication is hampered. Adding the effector molecule (pictured in dark blue) causes the building blocks responsible for effector recognition to connect to each other and produce a host that binds the effector. The removal of the effector molecules from the mixture results in a dramatic reduction of the diversity of the molecular network, and allows the self-replicator to form and grow.
The ability to trigger self-replication is an important step towards integrating replication with other key features of life. Such integration paves the way to constructing supersystems in which replication is controllable, and could bring the de novo synthesis of life a step closer.
- Effector-Triggered Self-Replication in Coupled Subsystems,
Dávid Komáromy, Meniz Tezcan, Gaël Schaeffer, Ivana Marić, Sijbren Otto,
Angew. Chem. Int. Ed. 2017.
DOI: 10.1002/anie.201707191