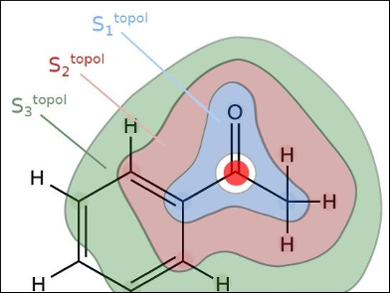
Cytochromes P450 (CYPs) are a large family of enzymes responsible for breaking down a wide array of compounds to which our bodies can be exposed, including drug molecules. The ability to predict the various sites of metabolism (SoM) on a given compound would be a significant benefit to researchers in drug design. This would allow researchers to fine-tune how quickly a drug is metabolized by CYPs and thus cleared from the body.
Gisbert Schneider, Swiss Federal Institute of Technology (ETH) Zurich, Switzerland, and colleagues have made it easier to predict a compound’s SoM. The method combines machine learning and a novel molecular representation that characterizes local atom environments using partial charge distributions derived from quantum-chemical calculations. The approach has a low computational cost.
The developed method is able to accurately identify SoM “soft spots” in drug molecules and serves as an alternative to time-consuming experimental metabolism studies. The researchers demonstrated the ability of their approach to rapidly identify the correct SoM in drugs with over 90 % accuracy. The team corroborated the predictions with experimental results for 25 drugs that are currently under pharmaceutical development.
- Site of Metabolism Prediction Based on ab initio Derived Atom Representations,
Arndt R. Finkelmann, Andreas H. Göller, Gisbert Schneider,
ChemMedChem 2017.
DOI: 10.1002/cmdc.201700097