Eighty years after Fleming’s Nobel Prize and the distribution of penicillin, antimicrobial resistance is one of the world’s top health priorities. The emergence of drug-resistant pathogens is jeopardizing modern medicine.
Corinne Lionne, Centre de Biologie Structurale (CBS), ABCIS Team, Montpellier, France, and Elise Kaplan, Unité de Microbiologie Moléculaire et Biochimie Structurale (MMSB), ATRA Team, Lyon, France, discuss their recent work, published with colleagues, identifying a new family of inhibitors that reverse aminoglycoside resistance in a strain of Enterococcus. Enterococcus can cause serious infections such as urinary tract infections, bloodstream infections, heart valve infections, gut inflammation, meningitis, and abdominal infections.
What did you do?
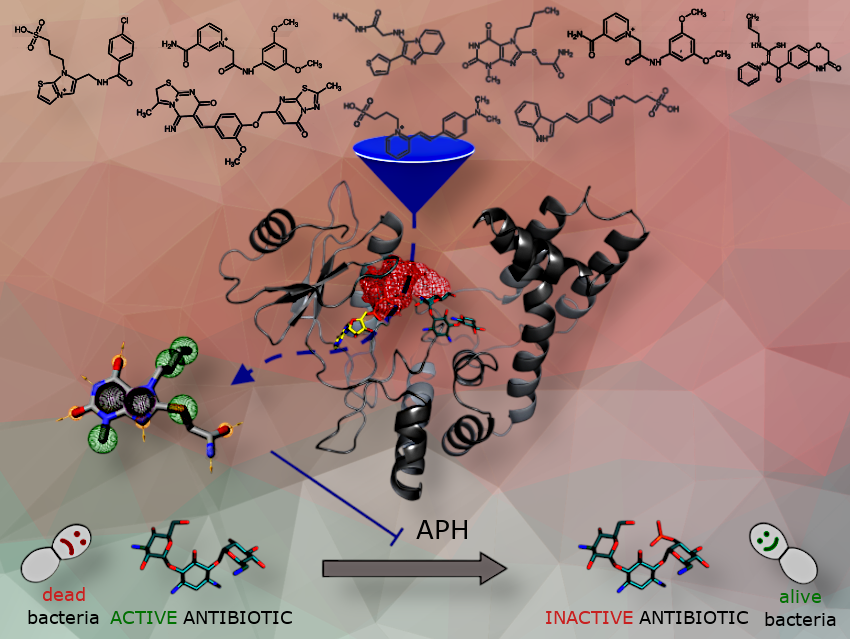
Resistance to aminoglycoside antibiotics is mainly due to small bacterial proteins, such as aminoglycoside-phosphotransferases (APHs), which inactivate the antibiotics. In our research article, we identified a new family of inhibitors that reverse resistance to aminoglycosides in a strain of Enterococcus.
Rather than experimentally screening a large number of molecules, we used computational methods to select candidate inhibitors from a chemical library. We performed molecular dynamics simulations and virtual screening on three APHs, targeting a druggable pocket conserved across all three proteins but distinct from their natural substrate binding sites.
The ability of the 14 top-ranked compounds to inhibit protein activity was then evaluated experimentally. Among these, the compound called EK3 non-competitively inhibited an APH, and its efficiency was further increased by slightly modifying the structure of the drug. These molecules restored the sensitivity of a resistant strain of Enterococcus to various aminoglycosides.
Why are you interested in this?
Antimicrobial resistance is a global concern, and an alarming low number of antibiotics have been introduced on the drug market in recent decades. One strategy is to find solutions that restore the effectiveness of existing antibiotics, whose use has been compromised by bacterial resistance.
What is new and cool about your work?
The high success rate of our screening approach is quite striking. We screened a small library of 12,000 molecules and identified a hit molecule by purchasing just 14 compounds. To our knowledge, this is the first allosteric inhibitor of an APH capable of restoring the sensibility of resistant bacteria.
In addition, this research also provides a proof-of-concept for the search for allosteric modulators using computational methods combined with experimental techniques.
What is the main significance of your results?
The study revealed that EK3 molecules were effective in restoring susceptibility to several aminoglycosides in resistant bacteria. The drugs were not toxic to human HeLa cells [cancer cell line], which is encouraging for their potential therapeutic use.
Although the precise location of the inhibitor within the APH is not known, enzymatic techniques made it possible to determine that the hit molecule does not compete with the enzyme natural substrates. This was a key objective as APHs share a nucleotide-binding site structurally related to eukaryotic kinases which are present in humans. It was therefore important to identify a molecule specific to APHs to avoid side effects on kinases when they are administrated.
What specific applications do you imagine?
We envision combined therapies where these drugs prevent the alteration of aminoglycosides, restoring the antibiotic’s ability to treat bacterial infections. The hit molecules will undergo further chemical optimization to improve their efficiency and advance to the next stages of preclinical research, specifically evaluating their effectiveness in combating bacterial infections.
What part of your work was the most challenging?
There were two challenging parts: The first was obtaining sufficiently stable and soluble molecules for the experimental tests, as a significant number of compounds were excluded from the enzymatic assays due to a lack of solubility.
The second challenge was to identify precisely the drug binding site in the APH enzyme to facilitate the rational design of more active compounds.
How can we tackle antibiotic resistance more effectively?
It is decisive to diversify our strategies for combating antibiotic resistance. We should seek promising molecules in chemical libraries and from natural sources. We need to combine high-throughput and virtual screenings, including AI-based tools. Fundamental research is also required to provide detailed insights into the diverse mechanisms behind bacterial resistance.
Thank you very much for sharing these insights.
The paper they talked about:
- APH Inhibitors that Reverse Aminoglycoside Resistance in Enterococcus casseliflavus,
Elise Kaplan, Laurent Chaloin, Jean-François Guichou, Kévin Berrou, Rahila Rahimova, Gilles Labesse, Corinne Lionne,
ChemMedChem 2024.
https://doi.org/10.1002/cmdc.202400842
Corinne Lionne is Research Director of the Centre de Biologie Structurale (CBS), ABCIS Team, in Montpellier, France.
Elise Kaplan is a Research Scientist at the Unit of Microbiologie Moléculaire et Biochimie Structurale (MMSB), ATRA Team, Lyon, France.