Resistance to antibiotics is a rapidly growing problem in health care. New targets to combat antibiotic resistance need to be identified and tackled as quickly as possible.
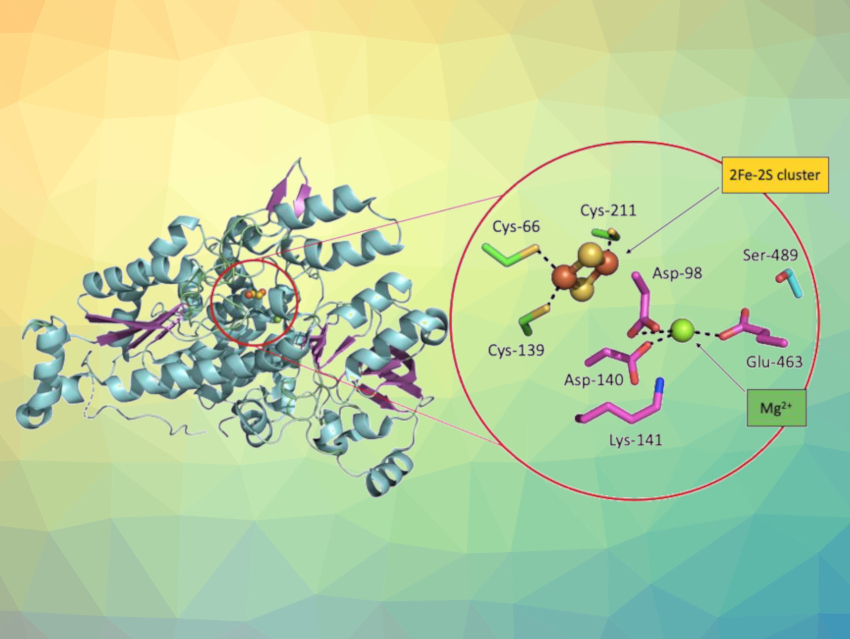
Gerhard Schenk, The University of Queensland, Brisbane, Australia, and colleagues have found that enzymes from the branched-chain amino acid biosynthesis pathway are attractive targets for the development of antimicrobial chemotherapeutics. The team produced two of these enzymes using E. coli bacteria, namely, Fe-S cluster-dependent dihydroxy-acid dehydratases (DHAD) from the human pathogens Staphylococcus aureus and Campylobacter jejuni (SaDHAD or CjDHAD, respectively). Both of these pathogens can show resistances to commonly used antibiotics. DHAD catalyzes, e.g., the dehydration of dihydroxy–isovalerate (DHIV) to keto–isovalerate (KIV).
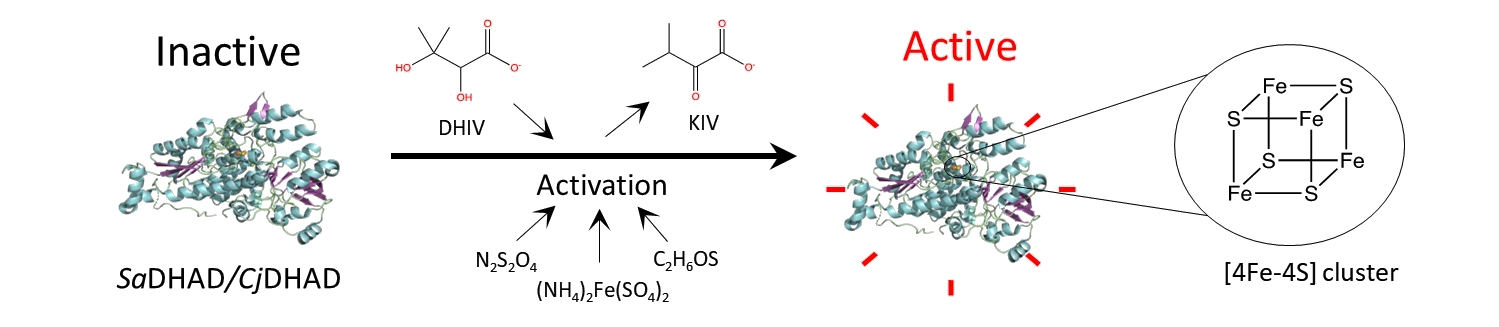
As isolated, the DHADs from these pathogens are inactive (pictured below on the left). However, the enzymes can be switched on by adding iron to form an [4Fe-4S] cluster in the active site (pictured below on the right). Although the activity gradually subsides, it can be boosted again, suggesting that the enzyme is structurally stable.
The team found that DHAD can be effectively inhibited by the natural product aspterric acid, a compound extracted from a fungus. This compound is, thus, a promising lead to develop much-needed new antibiotics.
- Dihydroxy‐acid dehydratases from pathogenic bacteria: emerging drug targets to combat antibiotic resistance,
Tenuun Bayaraa, Jose Gaete, Samuel Sutiono, Julia Kurz, Thierry Lonhienne, Jeffrey R. Harmer, Paul V. Bernhardt, Volker Sieber, Luke Guddat, Gerhard Schenk,
Chem. Eur. J. 2022.
https://doi.org/10.1002/chem.202200927