Measuring heat changes can be crucial for understanding interactions between molecules. However, it is very challenging to measure heats in small open systems. Such systems equilibrate quickly with their surroundings, mitigating any temperature changes.
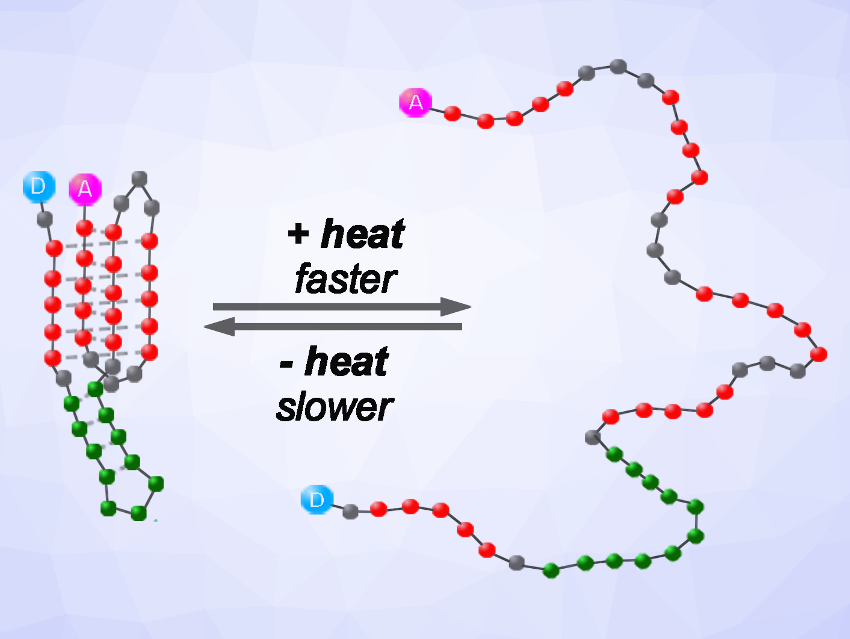
To tackle this challenge, Irina Nesterova and colleagues, Northern Illinois University, DeKalb, USA, have designed a molecule with “memory” that functions as a calorimeter. Nucleic acids are well known to fold or unfold in response to temperature variations. The team’s nucleic-acid-based sensor switches conformation in response to changes in heat and memorizes the response even after the change dissipates. It consists of a so-called DNA i-motif, i.e., a quadruplex structure formed from cytosine-rich single-stranded DNA. The molecule unfolds fast in response to heat, but is extremely slow to fold back (pictured). Changes in emission can be used to monitor this process.
As opposed to a thermometer, such memory-based sensors can assess heat changes regardless of whether the system equilibrates with its surroundings or not. Molecular calorimeters can be useful in a wide range of situations that are currently inaccessible, e.g., measuring heat conversions in very small volumes, providing high spatial resolution, or working in high-throughput formats. On a broader scale, memory-based molecular sensors could also be used to assess other small dissipating physical cues or transient quantities of analytes.
- Rational Design of Memory‐Based Sensors: the Case of Molecular Calorimeters,
Irina V. Nesterova, Obianuju A. Nwokolo, Brant Kidd, Te’Kara Allen, Alexander S. Minasyan, Suchitra Vardelly, Kristopher D. Johnson,
Angew. Chem. Int. Ed. 2020.
https://doi.org/10.1002/anie.202011422