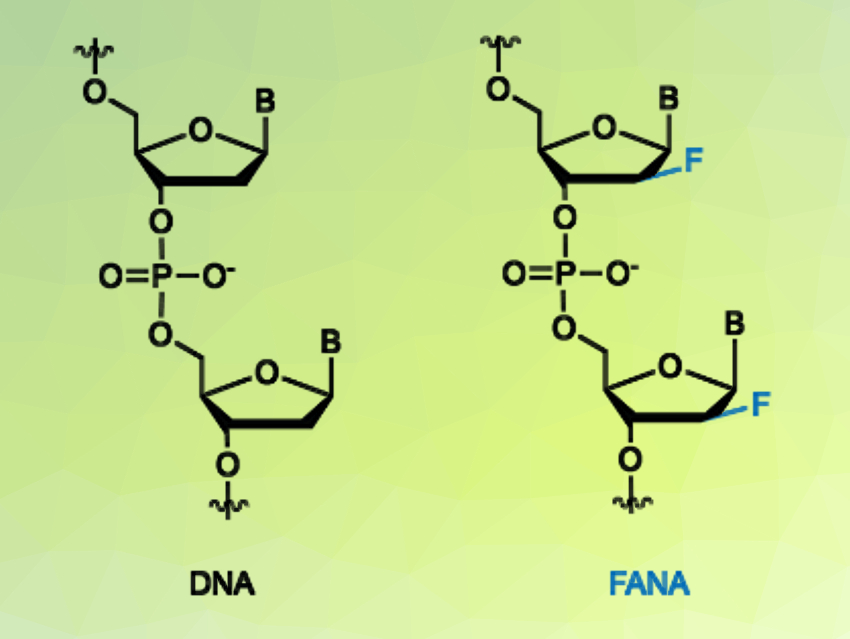
Artificial genetic polymers, also known as xeno-nucleic acids or XNAs, have unique physical and chemical properties that could allow them to catalyze chemical reactions with high efficiency. However, most of these XNA enzymes (XNAzymes) generated by in-vitro selection have a much lower activity than their “natural” DNA and RNA counterparts.
John C. Chaput, University of California, Irvine, USA, and colleagues have explored the catalytic potential of an XNAzyme that is composed entirely of 2′-fluoroarabino nucleic acid (FANA, pictured right), named FANAzyme 12-7. This enzyme was evolved to cleave RNA at a specific phosphodiester linkage. The team found that the FANAzyme outcompetes equivalent DNAzymes in model reactions performed under a range of conditions, including those that simulate the physiological environment of a cell.
According to the researchers, this is the first example of an in vitro-selected XNA enzyme with catalytic efficiencies greater than a DNAzyme. This might be important for the evolution of XNA catalysts and future applications in synthetic biology.
- Evaluating the Catalytic Potential of a General RNA‐Cleaving FANA Enzyme,
John C. Chaput, Yajun Wang, Alexander Vorperian, Mouhamad Shehabat,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900596