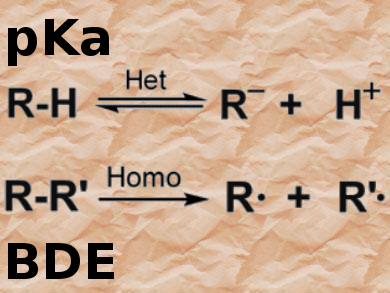
The importance of pKa values (Bronsted acidities) in rationalizing and organizing organic reactivity cannot be overestimated. Thanks to several valuable compilations, pKa values have been available online for many years. However, all of these online lists have limitations — for example, coverage of the current literature and/or only available for certain solvents. Therefore, the recently published (October 2016) database iBonD 2.0, which contains more than 30,000 pKa values for about 20,000 compounds in various solvents as well as 7500 homolytic bond-dissociation energies, can be considered the gold standard for accessing such data.
Under the direction of Professor Jin-Pei Cheng, a former student of the legendary Frederick G. Bordwell and active researcher in this field, a group of about 60 scientists and students from Nankai University, Tianjin, and Tsinghua University, Beijing, China, have created this magnum opus, which can be expected to become “the organic chemist’s best friend”.
iBonD 2.0
This database, which was presented at the IUPAC International Conference on Physical Organic Chemistry in Sydney in July 2016, is fascinating not only for its comprehensiveness, but also its user-friendly search methods. One can select a certain class of compounds to get an overview of the pKa range that they cover, or one can search for specific molecules, which can be entered by name, molecular formula, or structure by using the integrated structure editor.
The operation is best illustrated with a simple example. On entering the structure of pyrrolidine and selecting “substructure search”, 400 hits will be obtained, starting with the structure sought followed by that of the conjugate acid (pyrrolidinium ion), as shown in Table 1. The dissociated proton is shown in blue, which becomes important for molecules with various acidic sites (di- or polyacids).
Table 1. First hits of the substructure search in iBonD 2.0 for pyrrolidine.
The pKa values are shown for all solvents used, including gas-phase acidities, which are given as ΔG0acid in kilocalories per mole. The listing shows the method of determination (abbreviations are explained in an extra list in iBonD) as well as a hyperlinked code for the reference, which appears when the mouse pointer hovers over the link. Clicking the link will take the users to the full document.
The display for the homolytic bond dissociation energies (BDEs) is similar and gives BDE values in kilocalories per mole. For cases in which different BDEs have been published for the same molecule, the most recommended value is usually marked with a star.
Conclusion
Of course, one cannot expect an early version of such a huge database to be without errors. Users should therefore feel free to use the feedback link to inform the iBonD team of any corrections.
The database is provided free of charge for nonprofit academic use and only requires simple registration. Users are requested to cite the following website in their publications/presentations whenever applicable: ibond.chem.tsinghua.edu.cn or ibond.nankai.edu.cn. It is a matter of fairness to follow this request and thus acknowledge the great service of the iBonD team to the chemical community.
Herbert Mayr, Ludwig-Maximilians-Universität (LMU) München, Germany