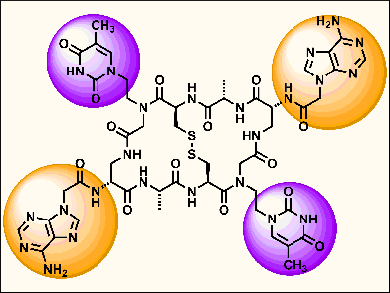
The cyclic peptide Triostin A, derived from Streptomyces triostinicus, is known to have potent antibacterial and antitumor activity because its quinoxaline side-chains can bisintercalate DNA. Triostin A can be modified to replace the quinoxalines with nucleobases that can be effective DNA major groove binders.
Ulf Diederichsen, Georg-August-University Göttingen, Germany, and colleagues synthesized Triostin A analogues with four nucleobase recognition units (ATTA or TTTT). The four recognition units were organized on a rigid template and were well-suited for DNA double strand interactions. These compounds, called aza-TANDEM derivatives, interacted with DNA double strands with the proper sequence complementarity and topological fitting. The purified compounds were then tested for self-aggregation and were shown to exhibit a median lethal dose (LD50) of >5 mg/mL in an Artemia salina assay.
- Triostin A Derived Cyclopeptide as Architectural Template for the Alignment of Four Recognition Units,
Ursula M. Kotyrba, Kevin Pröpper, Eike-F. Sachs, Anastasiya Myanovska, Tobias Joppe, Friederike Lissy, George M. Sheldrick, Konrad Koszinowski, Ulf Diederichsen,
ChemistryOpen 2014, 3, 152–160.
DOI: 10.1002/open.201400001