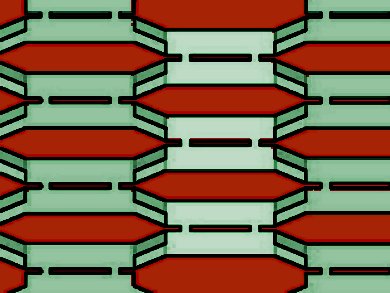
Macromolecular self-assembly is common in nature as a mechanism for the construction of complex structures. Crystals formed from DNA tiles are particularly interesting in their ability to self-assemble based on designed, single-stranded DNA “sticky ends” that attach to complementary ends on other tiles. This allows crystals to be designed that have complex, programmable structure despite consisting of only a relatively small number of different tile types.
While the theoretical implications of models of DNA tile self-assembly have been extensively researched and such models have been used to design DNA tile systems for use in experiments, there has been little research testing the fundamental assumptions of those models.
Erik Winfree and colleagues, California Institute of Technology, Pasadena, USA, use direct observation of individual tile attachments and detachments of two DNA tile systems on a mica surface imaged with an atomic force microscope (AFM) to compile statistics of tile attachments and detachments.
The single-tileresolution AFM movies of single DNA tile system crystals verify some key assumptions of the kinetic Tile Assembly Model (kTAM), at least for growth on a surface. The technique also raises the perspective of examining growth on a surface in general at a level that has not previously been possible.
- Direct Atomic Force Microscopy Observation of DNA Tile Crystal Growth at the Single-Molecule Level,
Constantine G. Evans, Rizal F. Hariadi, Erik Winfree,
J. Am. Chem. Soc. 2012.
DOI: 10.1021/ja301026z