Drug development starts with finding molecules that bind tightly to disease-causing proteins. Researchers struggle to predict which structures will work and, thus, rely on unbiased screening to identify tight-binding lead molecules. Therefore, the more molecules a screening includes, the greater the chance of identifying a possible drug candidate.
Macrocycles are found as structural motifs in many drugs in clinical use. Nearly all of them are natural products or close derivatives. Dennis Gillingham and colleagues, University of Basel, Switzerland, have created a diverse scaffold collection of synthetic macrocycles to find lead structures that can inhibit disease-relevant proteins.
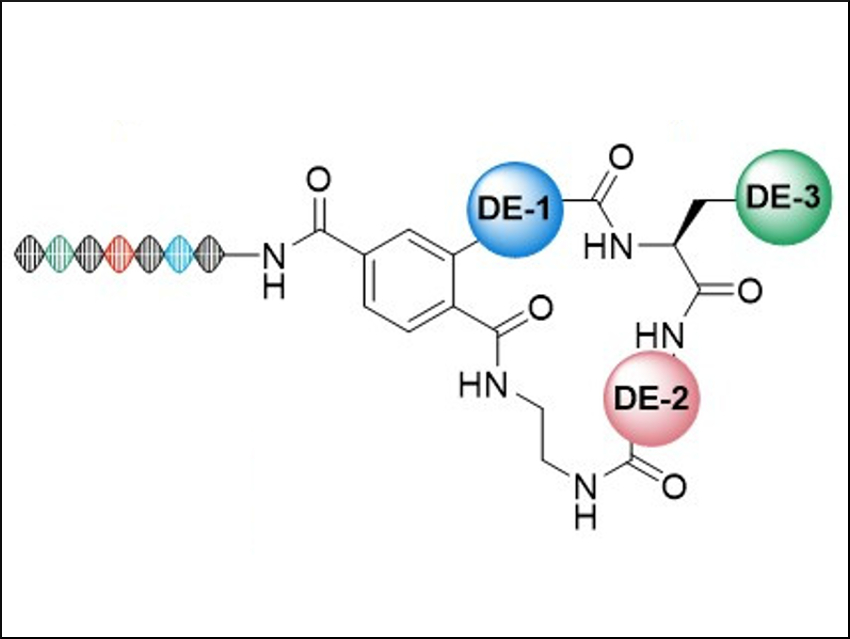
The macrocycles contain subunits that are similar to natural bioactive compounds (general structure pictured). To create the macrocycles, the team synthesized bifunctional precursors and used them in a combinatorial library synthesis. The library contains 2142 macrocyclic scaffolds, each with 663 side-chain variations—i.e., over a million candidates overall.
To keep track of molecular information, the team used “DNA barcodes” (pictured on the left). They allow to screen all library members simultaneously. This physical connection between DNA and drug candidate means that the tightest-binding compounds in a test can be determined by standard DNA sequencing.
- A DNA-encoded chemical library incorporating elements of natural macrocycles,
Cedric Stress, Basilius Sauter, Lukas Schneider, Timothy Sharpe, Dennis Gillingham,
Angew. Chem. Int. Ed. 2019.
https://doi.org/10.1002/anie.201902513