RNA probes are used to follow protein expression in living systems. Development of fluorescent RNA probes is of great significance in cell, molecular, and structural biology. One main challenge in RNA sensing is the lack of predictive ability for RNA secondary structures, which hinders the development of sensors that rely on conventional Watson and Crick base-pair interactions.
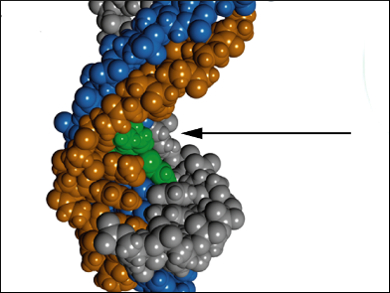
Seiichi Nishizawa, Tohoku University, Sendai, Japan, and colleagues optimized triplex-forming peptide nucleic acid (TFP) probes (pictured orange) containing thiazole orange (TO, pictured green) as a response element. The TFP probes interact with double-stranded RNAs (dsRNA, pictured blue/gray) through the unconventional Hoogsten base-pairing at pH 5.5. Upon interaction, TO intercalates into the triplex structure and restricts its movement. This restriction results in an increase in TO fluorescence intensity.
The team tested different linkers between the TFP backbone and TO and found that a propyl linker is the optimal linker for dsRNA sensing. They also showed that TO interacts with any base in the target sequence, making it a universal base. However, mismatches between the probe and the target in other regions of the sequence resulted in a lower TO fluorescence intensity. This effect indicates specificity of the probe’s light-up response, which makes it ideal for RNA sensing applications.
- Optimization of the Alkyl Linker of TO Base Surrogate in Triplex-Forming PNA for Enhanced Binding to Double-Stranded RNA,
Takaya Sato, Yusuke Sato, Seiichi Nishizawa,
Chem. Eur. J. 2017.
DOI: 10.1002/chem.201604676